
IntroductionĬOnstraint-Based Reconstruction and Analysis (COBRA) methods have been successfully employed in the field of microbial metabolic engineering (1-3) and are being extended to modeling transcriptional (4-8) and signaling (9-11) networks and the field of public health (12). This Toolbox lowers the barrier of entry to use powerful COBRA methods. A suite of test scripts can now be used to learn the core functionality of the Toolbox and validate results. In version 2.0, we improved performance, usability, and the level of documentation.

As with the first version, the COBRA Toolbox reads and writes Systems Biology Markup Language formatted models. New functions include: (1) network gap filling, (2) 13C analysis, (3) metabolic engineering, (4) omics-guided analysis, and (5) visualization. Version 2.0 of the COBRA Toolbox expands the scope of computations by including in silico analysis methods developed since its original release.
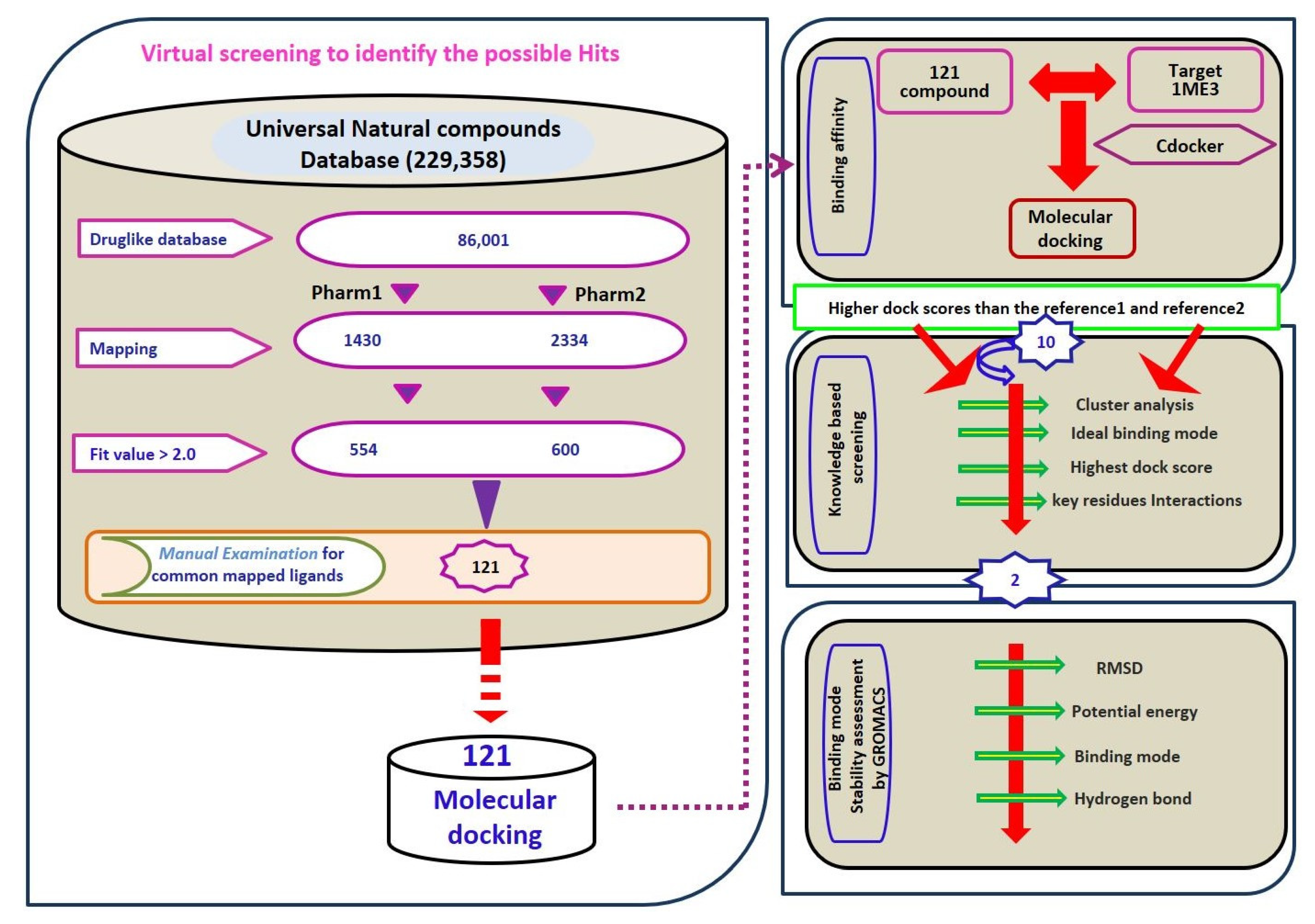
UNIVERSAL DATABASE METABOLIC REACTIONS UPDATE
Here we present a significant update of this in silico ToolBox. The COBRA Toolbox, a MATLAB package for implementing COBRA methods, was presented earlier. Over the past decade, a growing community of researchers has emerged around the use of COnstraint-Based Reconstruction and Analysis (COBRA) methods to simulate, analyze and predict a variety of metabolic phenotypes using genome-scale models. Authors: Daniel Hyduke, Jan Schellenberger, Richard Que, Ronan Fleming, Ines Thiele, Jeffery Orth, Adam Feist, Daniel Zielinski, Aarash Bordbar, Nathan Lewis, Sorena Rahmanian, Joseph Kang & Bernhard Palsson


 0 kommentar(er)
0 kommentar(er)
